Researchers record the first "film" of the multiplication of the influenza A virus genome
11.09.2024
 |
|
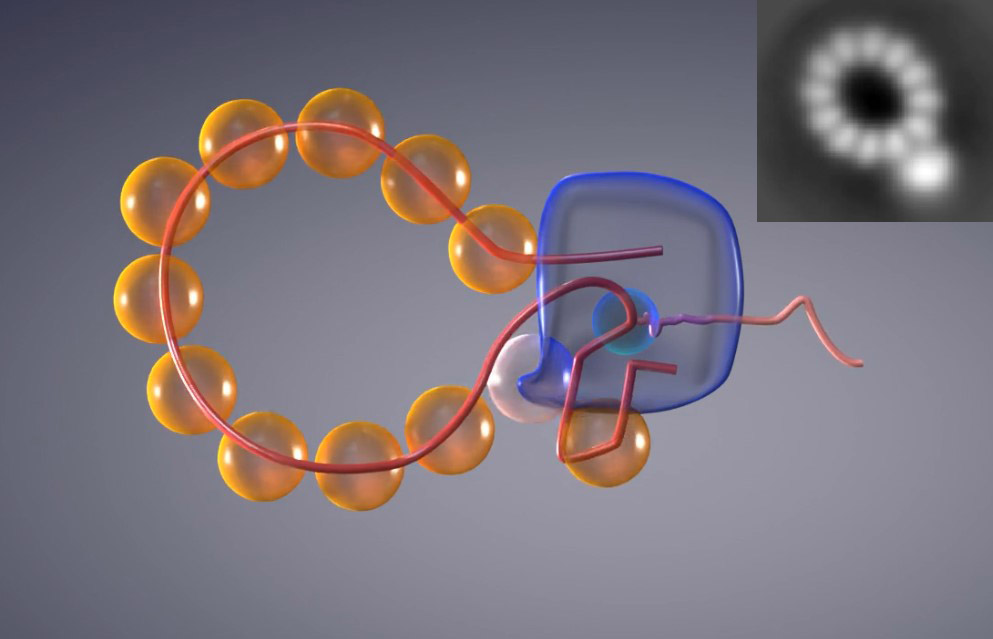
Recombinant genome of influenza A virus, during the transcription process. Credit: Patricia Bondía, Enrique Sahagún (Scixel). |
- IMDEA Nanociencia researchers observe live, for the first time, the multiplication of the influenza A virus genome.
- The "films" obtained allow us to understand some of the factors that determine the rate of multiplication of the influenza virus.
| Tweet |
Madrid, 11th September, 2024.
The influenza A virus is a major threat that concerns the public health. Understanding how this virus replicates is crucial, especially given that its mutations can give rise to new strains capable of affecting humans. The nucleus of the virus encloses its genetic information, contained in RNA chains – ribonucleic acid – which the polymerase enzyme copies to generate new viruses. RNA strands are covered by proteins that protect RNA from being degraded inside cells. How does polymerase manage to multiply RNA efficiently if it is completely covered by proteins? And besides, how does it manage to copy RNA without decoupling it from the proteins that protect it?
During the process of RNA multiplication, the viral polymerase moves through the RNA structure, synthesizing and copying the structure. The proteins that protect the RNA of the influenza A virus genome are arranged in the form of a compact double helix, masking the position of the polymerase. Thus the polymerase in action cannot be observed and many details of the "copying" process are hidden. To date, it has not been possible to track the movement and activity of the polymerase throughout the genome of the virus.
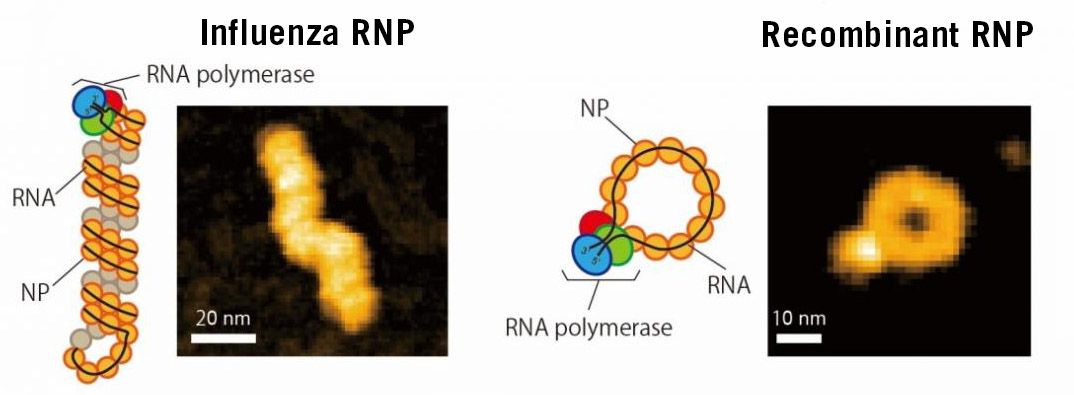 The "Manipulation of Molecular Motors" research group of the IMDEA Nanociencia institute (Madrid), led by Borja Ibarra, in collaboration with researchers from NanoLSI (Kanazawa University Japan) and the National Center for Biotechnology (CNB-CSIC Madrid) have devised a strategy that is key to studying this elusive process in detail. The researchers shortened the virus genome so that the proteins form a ring rather than an helix. In this way, the position of the polymerase is exposed.
The "Manipulation of Molecular Motors" research group of the IMDEA Nanociencia institute (Madrid), led by Borja Ibarra, in collaboration with researchers from NanoLSI (Kanazawa University Japan) and the National Center for Biotechnology (CNB-CSIC Madrid) have devised a strategy that is key to studying this elusive process in detail. The researchers shortened the virus genome so that the proteins form a ring rather than an helix. In this way, the position of the polymerase is exposed.
With this strategy, the researchers were able to analyze the motion of the polymerase in real-time, using high-speed atomic force microscopy. They recorded multiple "live" films of the process which, combined with electron microscopy images, helped them understand and unveil novel information about the molecular processes that govern the amplification of the viral genome. The researchers observed that the polymerase manages to access RNA without separating it from the proteins that protect it. This is essential because it preserves the structure of the genome, which in turn, allows it to multiply continuously. The polymerase is capable of producing multiple copies from the same parent RNA in several rounds, which is a key aspect for viral multiplication.
A control mechanism in viral multiplication
These nanoscopic "films" allowed the researchers to estimate the rate of RNA synthesis, the speed at which the viral polymerase works. The polymerase is capable of incorporating up to 35 nucleotides in one second. If we equate a nucleotide with a letter, a copyist working at this speed would be able to copy the first part of Don Quixote in just 6 hours (or the first Harry Potter book in 3 hours).
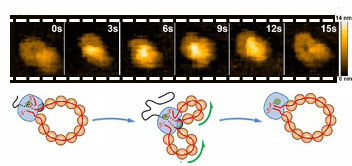 The team of researchers also discovered that nascent RNA, which can restructure and fold, influences the rate at which the polymerase works. The conformation of the nascent RNA therefore functions as a control mechanism that regulates the rate of amplification of the virus and could be a therapeutic target for the development of new antiviral strategies.
The team of researchers also discovered that nascent RNA, which can restructure and fold, influences the rate at which the polymerase works. The conformation of the nascent RNA therefore functions as a control mechanism that regulates the rate of amplification of the virus and could be a therapeutic target for the development of new antiviral strategies.
The context does matter
Viral RNA multiplies itself surrounded by proteins. Unlike previous work that studied the isolated polymerase, this finding has been made in the natural environment of the polymerase, inside the genome and surrounded by proteins that it has to deal with and that affect the final amplification rate. The model system of this study provides direct evidence that individual viral proteins can be recycled, and confirms existing theoretical models. The work has been well received by the scientific community and offers a new approach to investigate the mechanisms of viral transcription and replication in other viruses.
When Borja Ibarra is asked about the importance of knowing in detail the functioning of the virus's polymerase, he answers: "If we manage to define the mechanisms that govern the functioning of viral proteins, we will be able to devise methods to interfere with them and, therefore, stop the viral infection". The work, recently published in ACS Nano, lays the foundations for future research on the functioning of the polymerase in the context of the viral genome, something not possible until now.
This work has been carried out at IMDEA Nanociencia, the National Center for Biotechnology (CSIC) and the Nano Life Science Institute of Kanazawa University and is partially funded by the Severo Ochoa Seal of Excellence awarded to IMDEA Nanociencia (CEX2020-001039-S). The work has received support from the 'NanoLSI Visiting Fellows' grant program awarded to Borja Ibarra, which funds the stay of 3 researchers each year for 12 months at the Japanese institute.
Glossary:
Polymerase: enzyme responsible for the replication or transcription of nucleic acids, (DNA or RNA).
RNA: a macromolecule formed by a single chain of nucleotides that participates in essential biological processes for living beings together with DNA.
Nucleotide: the basic fundamental molecule of nucleic acids (RNA and DNA).
Reference:
Diego Carlero, Shingo Fukuda, Rebeca Bocanegra, Toshio Ando, Jaime Martin-Benito and Borja Ibarra. Conformational dynamics of influenza A virus ribonucleoprotein complexes during RNA synthesis. ACS Nano. 18, 19518 (2024). https://doi.org/10.1021/acsnano.4c01362
![]() https://repositorio.imdeananociencia.org/handle/20.500.12614/3750
https://repositorio.imdeananociencia.org/handle/20.500.12614/3750
Contact:
Dr. Borja Ibarra
This email address is being protected from spambots. You need JavaScript enabled to view it.
Molecular Motors Manipulation Group
https://nanociencia.imdea.org/molecular-motors-manipulations-lab/group-home
Twitter: @3m_lab
Oficina de Divulgación y Comunicación en IMDEA Nanociencia
divulgacion.nanociencia [at]imdea.org
Twitter: @imdea_nano
Facebook: @imdeananociencia
Instagram: @imdeananociencia
Source: IMDEA Nanociencia.
IMDEA Nanociencia Institute is a young interdisciplinary research Centre in Madrid (Spain) dedicated to the exploration of nanoscience and the development of applications of nanotechnology in connection with innovative industries.